To encourage participants from non-medical domains (who don’t have clinical data available) we are providing real multi-modal clinical data. To access the data, please send your request to cerevis2018@gmail.com.
In your request, please specify your name, job/title, institute, work address, name of the dataset, and a brief but specific description of how you would intend to use the data. By requesting the data, the authors imply their intention to submit to CereVis2018 workshop.
Privacy and data copyright
By requesting the data, the authors agree to use the provided data only in the scope of the CereVis workshop and neither pass it onto a third party nor use it for other publications. No copyright transfer of any kind will take place, except in the case of a contribution to the LNCS proceedings of the workshop.
The demo images shown on this website are not permitted to be used without the consent of the authors.
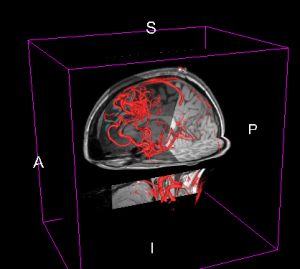
Brain arteriovenous malformation (AVM)
AVMs are abnormal collections of blood vessels in the brain. The central part of the AVM, the nidus, is made up of abnormal vessels that are hybrids between true arteries and veins. AVMs are fed by one or more feeding arteries (feeders) and are drained by one or more major draining veins (drainers). These feeding and draining vessels often have weakened walls and therefore may leak or rupture.
Neurosurgery for AVMs involves identifying the margins of the malformation and tying off or clipping the feeder vessels, obliterating the draining veins and removing or obliterating the nidus, in this order. Therefore, a detailed understanding of the arterial inflow from feeders and venous drainage from drainers is important for clinical evaluation and management of AVMs. The use of techniques that aid in the characterization of the pattern and distribution of feeding arteries by quantification of the relative blood flow is necessary as it is not always easy to identify whether a vessel is a feeding artery or an arterialized draining vein.
Data provided:
MR angiography, T1w & T2FLAIR MRI, and 3D tracked Doppler ultrasound in NIfTI format.
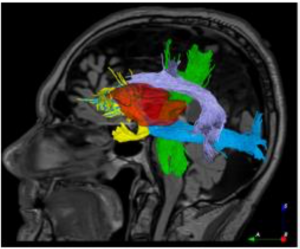
Brain tumor resection
In brain tumor surgeries, removing the maximum amount of cancerous tissue while preserving brain function can help improve survival and quality of life. Diffusion MRI is gaining growing significance in brain tumor resection thanks to its ability to reveal brain connections that helps optimize surgical plans while minimizing neurological deficits. Appropriate visualization of the spatial relation between the general brain anatomy, tumor, and important white matter fibre bundles computed from diffusion MRI can enable more effective craniotomy planning and decisions on the optimal path for resection.
Data provided:
T1w & T2FLAIR MRI for general anatomy, raw diffusion MRI, tumor segmentation, and processed tractography images. All images are available in NIfTI format, and tractography files are also available in .vtk format.
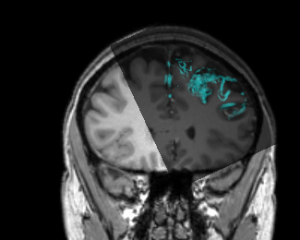
Stereo-Electroencephalography (SEEG) for Investigating Epilepsy
Patients with epilepsy may be investigated with intracranial electrodes to help identify the seizure focus. In a typical SEEG investigation, multiple electrodes are implanted (often 10-15 electrodes each with 5-10 electrode contacts each); thus resulting in over 100 contacts for electrophysiological recording. Visualizing the spatial relationship between individual electrode contacts can be challenging but is crucial for appropriate identification of the focus as the from these investigations may permit surgical resection or ablation of the focus with intent to cure.
Data provided:
A contrast-enhanced T1w image, post-operative computed tomography (CT) scan, electrode segmentation, and csv file with individual electrode contact coordinates is provided. A basic Slicer scene integrating all these modalities is also provided for reference. All images are available in NIfTI format, and surface model files are available in .vtk format.
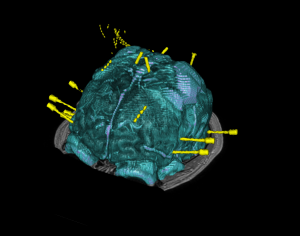
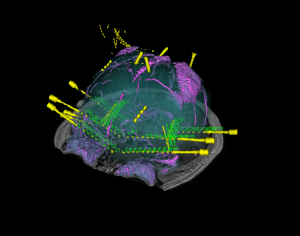
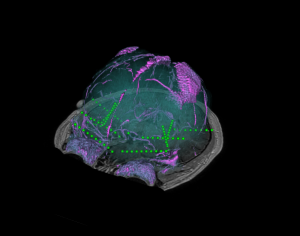
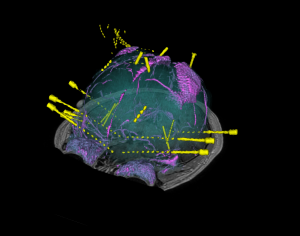
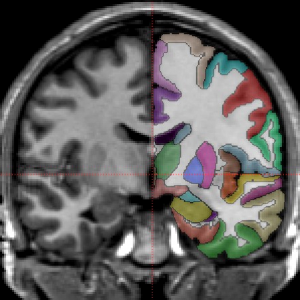
Brain cortical and subcortical segmentation
The human brain is a complex organ with numerous functional sub-structures. In medical education, brain research, and neurosurgeries, an easy-to-understand visualization of the brain structures and their spatial relationships is highly instrumental. However, it’s often not easy to show complex anatomy in 3D.
Data provided:
MRI scan and associated brain structural segmentation in NIfTI format. Labeled volumes have a number at each voxel that indicates which anatomical regions it belongs to. In the image below, the raw MRI signal for one slice is shown with slightly transparent labels on the right side.